Minimum Set Primers and Unique Probes Design Algorithms for
Differential Detection of Symptom-Related Pathogens
| HOME | |
| Introduction | |
| Methodology | |
| Tetra-Nucleotide Nucleation (TNN) | |
| Unique & Common Sections | |
| Nearest-Neighbor Model | |
| MCGA | |
| Linker Design | |
| Computational Results | |
| Bio-Experiment | |
| Conclusion | |
| Reference | |
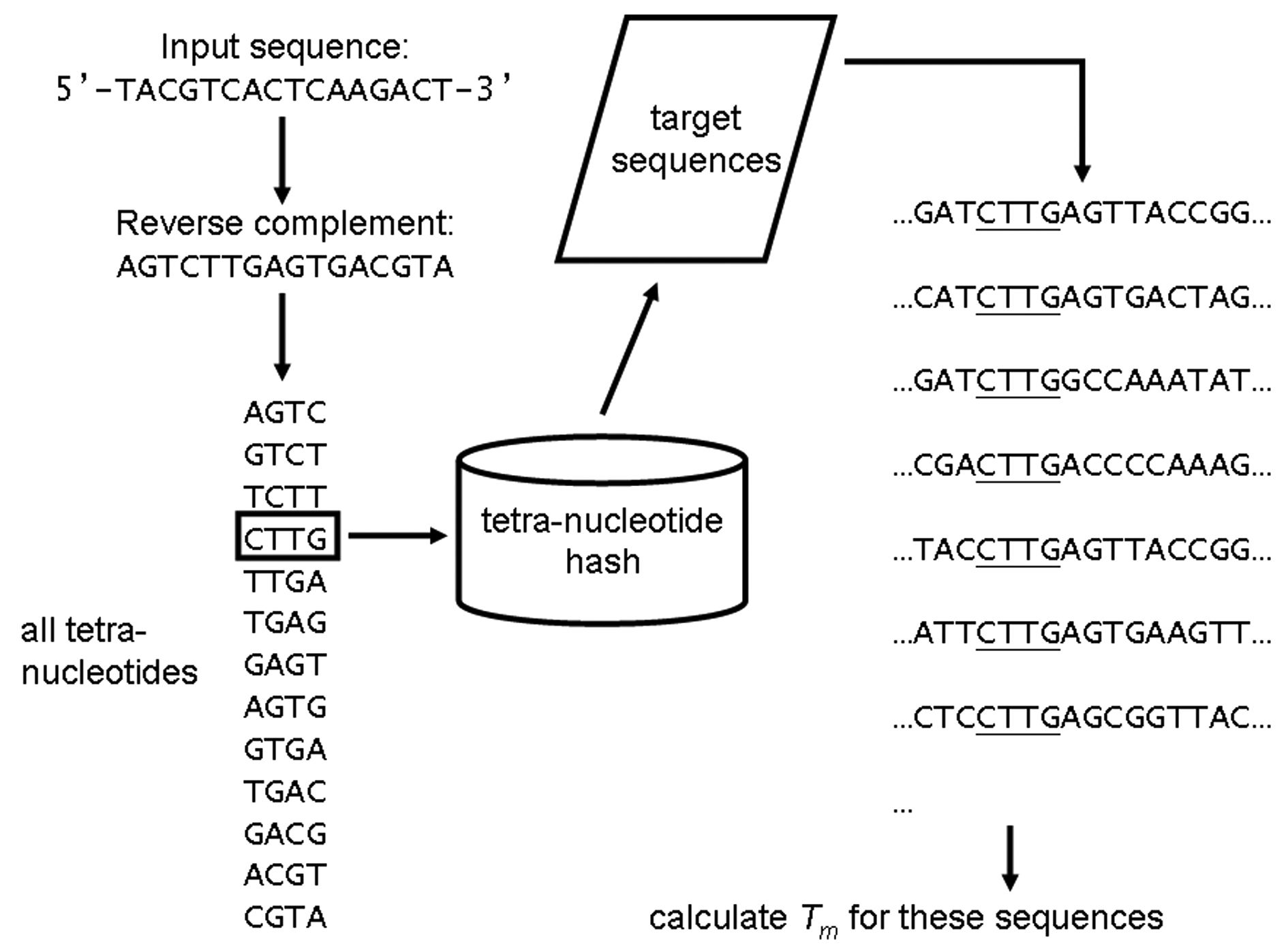
Fig. 2. Fast filtering of probes/primers with TNN hash. For target sequences, we built tetra-nucleotide nucleation (TNN) hash for efficient identification of unique and common sections. The hash contains the occurrences of each possible tetra-nucleotide and their respective locations on the target sequences. Based on the nucleation theory of primer/probe annealing, we have assumed that unique regions of a target sequence contain tetra-nucleotides with the least occurrences; and common sections contain tetra-nucleotides with the most counts. That is, a region that shares the least tetra-nucleotides with other sequences is likely to be unique. With an input sequence, the reverse complement of the sequence was generated and looked up in the hash table. For each tetra-nucleotide, the melting temperature ( T m ) of the sequence around each occurrences is calculated. These T m values are used to fast filter sequences which will (will not) be annealed to the input sequence. The process is illustrated in Fig. 2. A set of candidate sequences are searched based on entries in the TNN hash, and these sequences are screened with their melting temperature against the original query sequences. TNN hash is used through out the entire design process. Primers and linkers are all screened against the TNN hash. With the TNN hash, candidate sequences for primers and probes can be screened and evaluated rapidly. |