Minimum Set Primers and Unique Probes Design Algorithms for
Differential Detection of Symptom-Related Pathogens
| HOME | |
| Introduction | |
| Methodology | |
| Tetra-Nucleotide Nucleation (TNN) | |
| Unique & Common Sections | |
| Nearest-Neighbor Model | |
| MCGA | |
| Linker Design | |
| Computational Results | |
| Bio-Experiment | |
| Conclusion | |
| Reference | |
The thermodynamic parameters for hybridization between oligo-nucleotides and target sequences are estimated with a nearest-neighbor model. These thermodynamic parameters include melting temperatures ( Tm ), free energy ( Delta G ), enthalpy ( Delta H ), and entropy ( Delta S ). The nearest-neighbor model has been proven to accurately estimate thermodynamic parameters (Rahmann and Grafe, 2004; Tanaka, et al., 2004) . In the nearest-neighbor model, the thermodynamic parameters are calculated from hybridization of consecutive di-nucleotides. For example, the enthalpy will be calculated as follows:
where Delta Hnn is the enthalpy of hybridization between two di-nucleotides, and Delta Hinit is the enthalpy for initiation of a DNA duplex. The other thermodynamic parameters (free energy and entropy) are estimated in similar ways. The melting temperature ( Tm ) of annealed sequences are calculated as follows (Sugimoto, et al., 1996) :
|
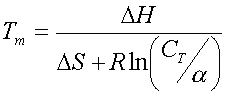 (5)
(5)