Minimum Set Primers and Unique Probes Design Algorithms for
Differential Detection of Symptom-Related Pathogens
| HOME | |
| Introduction | |
| Methodology | |
| Computational Results | |
| Bio-Experiment | |
| Conclusion | |
| Reference |
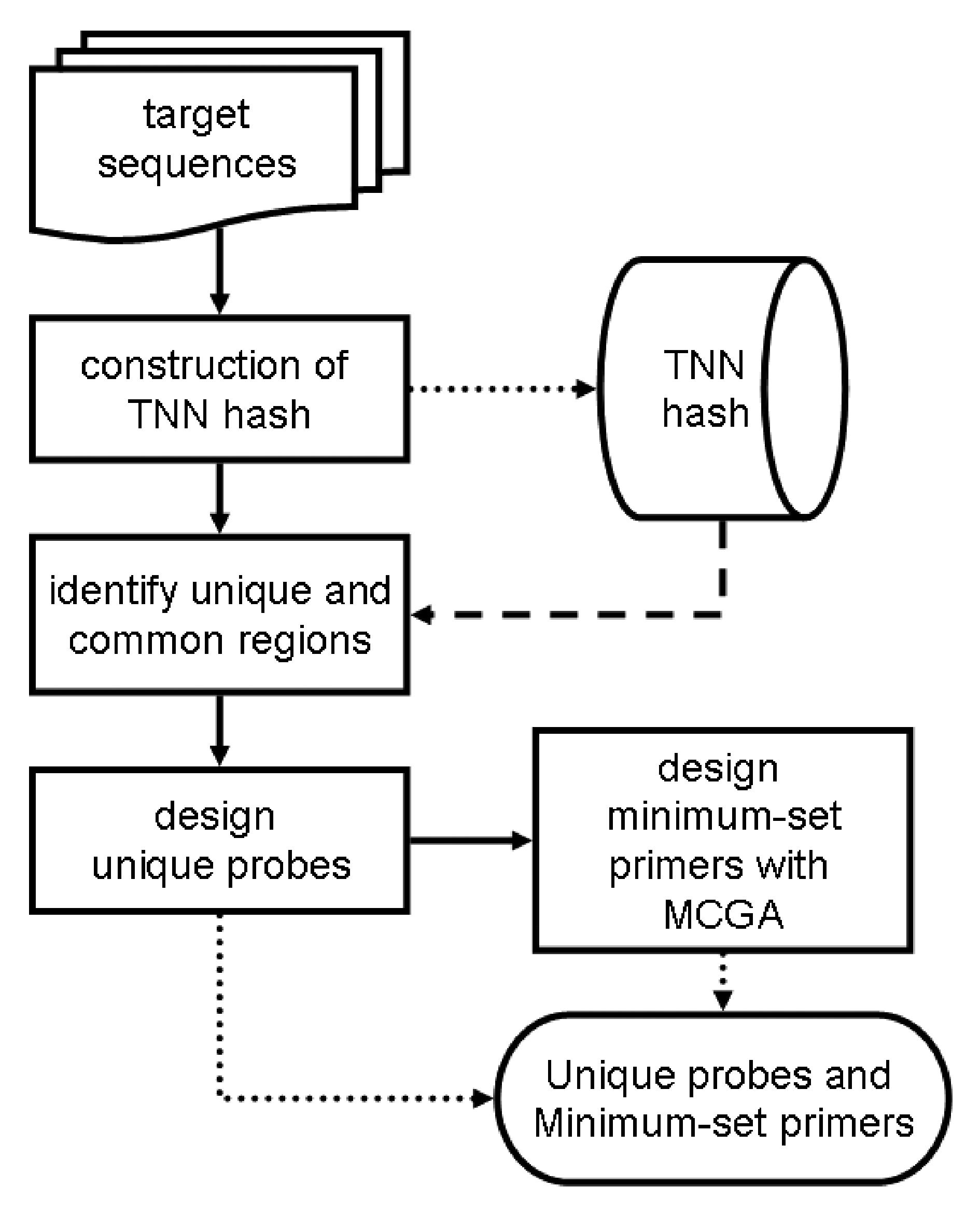
Fig.1. The overall flow of our integrated probe/primer design algorithm. The design of DNA probes aids the diagnosis of infectious diseases. The goal of diagnostic microbiology is to provide rapid and accurate detection of specific pathogens within a clinical specimen in an appropriate time. Genes or genomic sequences representing a specific group of infectious agents or a particular strain are selected. The sequences are then synthesized or cloned as probes to detect the presence of the specific agents through hybridization. Unique probes can assure the accurate identification of a specific pathogen. However, the amounts of DNA or RNA within the clinical specimen are rather small. To improve the sensitivity of pathogen detection, DNA amplification techniques like PCR are used. Conventional PCR primers are unique, therefore one pair of primers will selectively amplify one single gene. In the case of pathogen detection, amplifications of several genes from dozens of infectious agents are necessary. Clinical samples usually contain genetic materials from various organisms (including the host). A large number of primers is required in conventional PCR to amplify target sequences specifically and sensitively. However, a large number of unique primers implies high synthesis cost and has limited practical use of diagnostic PCR. It has been proposed that with carefully designed multiple-use primers, the number of primers will be reduced significantly (Fernandes and Skiena, 2002) . The concept of multiple-use primers can also be extended to work across several organisms. Selected gene sequences from several pathogen genomes can be amplified with multiple-use primers (Fernandes and Skiena, 2002) . A large number of target sequences from pathogens can be amplified and detected simultaneously. These target sequences will be amplified with a minimum set of primers. The presence of sequences from infectious pathogens will then be detected with unique probes. The multiple-use primers will increase the sensitivity of diagnostic PCR, while unique probes will increase the specificity. With multiple-use primers, the number of primers required to amplify a large number of sequences can be reduced. Current approaches use heuristic algorithms to reduce the number of primers (Fernandes and Skiena, 2002) , but either the process is time-consuming, or the solution quality is not satisfactory. In this paper, we propose a new algorithm to design minimum set primers and unique probes. The flow of our algorithm is shown in Fig 1. We aimed to provide optimum reduction of primers within a reasonable amount of time. With a carefully designed algorithm, unique and common sections on a set of target sequences can be identified. We extended the nucleation theory for primer annealing reaction with a Tetra-Nucleotide Nucleation (TNN) hash. Combined with several other criteria, TNN served as an index for fast identification of unique and common sections on target sequences. The design of unique probes and multiple use primers are base d on these sections. We treated the multiple - use primer design problem as a minimum set covering problem, and used modified compact genetic algorithm (MCGA) to solve it . Our algorithm significantly reduce s the number of primers required to amplify a large number of sequences compar ed to other approaches. Also the time required for the primer design is significantly shorter. The efficiency of our algorithm has been verified with a simulation on 12,669 sequences. This algorithm has been applied to differential detection of 9 plant viruses. |