Minimum Set Primers and Unique Probes Design Algorithms for
Differential Detection of Symptom-Related Pathogens
| HOME | |
| Introduction | |
| Methodology | |
| Tetra-Nucleotide Nucleation (TNN) | |
| Unique & Common Sections | |
| Nearest-Neighbor Model | |
| MCGA | |
| Linker Design | |
| Computational Results | |
| Bio-Experiment | |
| Conclusion | |
| Reference | |
The melting temperatures ( Tm ) of designed primers may have large variances. This usually causes problems in PCR experiments. To make sure the Tm of all designed primers are within a reasonable range, we have adopted a two phase primer design approach. In our approach, we first design a minimum set of 12-mer primers using MCGA. Additional 8-mer linkers linked to the 5' -end of these primers are designed subsequently. These linkers do not interfere with the annealing of original PCR templates. The sole purpose of these linkers is to equalize the melting temperatures of the final primers. The upstream sequences of the primers are filtered with the TNN hash to exclude linkers similar to target sequences and designed primers. The constraints on the selection of linkers can be illustrated with the following:
where L is the set of candidate linkers, T is the set of target sequences, and P is the minimum set of 12-mer primers. Also
where PL is the set of primers with linkers (20-mer primers). For each primer, a linker will be selected so the melting temperatures of the primers with linkers will stay in a narrow range around 55°C . We have employed dual-phase PCR to validate the applicability of these primers with linkers. |
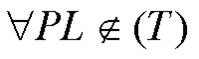 (11)
(11)