Minimum Set Primers and Unique Probes Design Algorithms for
Differential Detection of Symptom-Related Pathogens
| HOME | |
| Introduction | |
| Methodology | |
| Computational Results | |
| Bio-Experiment | |
| 9 Plant Virus (Table 3) | |
Primers & Probes (Table 4, 5) |
|
| Experimental Result | |
| Conclusion | |
| Reference | |
|
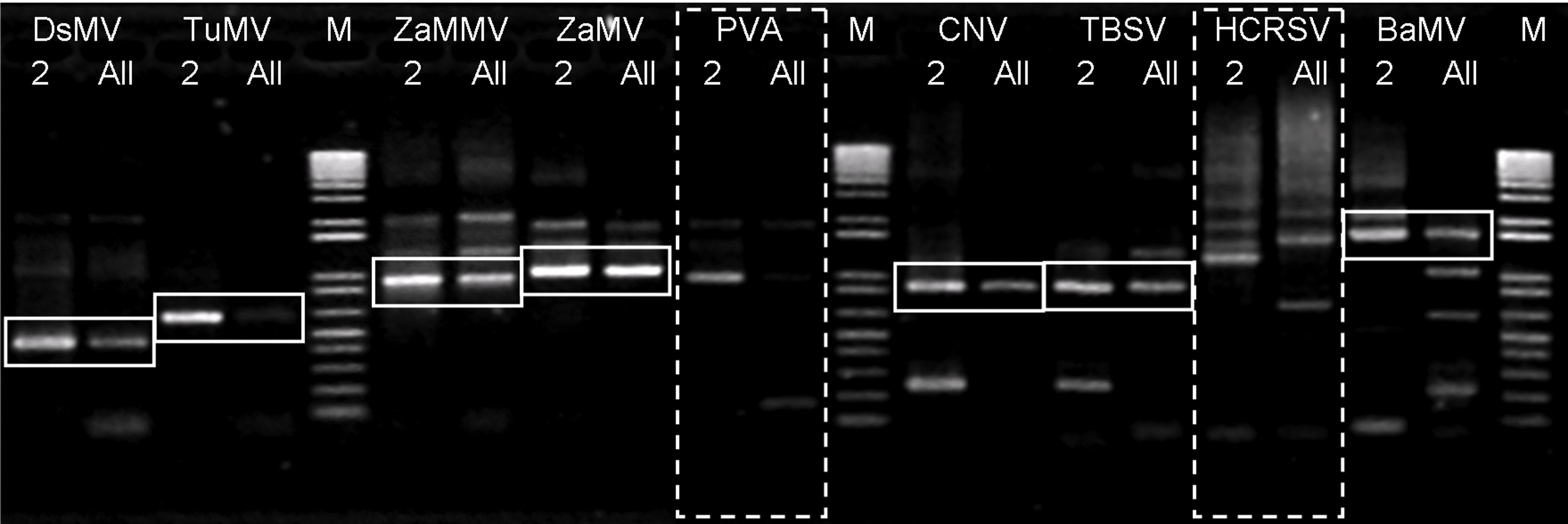
Fig. 3. The gel electrophoresis experiment for the verification of the designed primers. The lanes with label ‘M' are markers. Each virus has two lanes. The lanes with label ‘ 2' is the target sequence amplified with only its specific primers present. The lanes labeled ‘All' is the target sequence amplified with all 11 primers present.
Fig. 4 Triplicate dot hybridization with specific probes for each of the 9 plant viruses. The relative positions of probes for each target sequences are illustrated in (A). For brevity, only three slides are shown here. It is clear that (B) PVA, (C) TuMV, and (D) HCRSV can be detected with their respective unique probes.
Our minimum set primer design algorithm has obtained very good results with simulation on genome-wide PCR primer design. In this section we will combine our unique probe and minimum set primer design algorithm. The overall design algorithm will be applied to differential detection of 9 plant viruses from 4 genus. The design of our probes/primers are verified with wet lab PCR experiments. The 9 plant viruses are listed in Table 3. In conventional PCR, 18 primers will be necessary for amplification of 9 sequences. We have designed 11 primers from 11,067 candidates. These 11 primers cover the 9 target sequences. The reduction rate of the primer number is 39%. The cost for synthesis these primers is also reduced. These primers are listed in Table 4. The sequences of these primers and their respective Tm are provided. Except for primers LMu-01, LMu-02, LMu-05, and LMu-08, all primers are multiple-use primers and shared by more than one sequences. For example, LMu-04 is the forward primer for Bamboo mosaic virus (BaMV) and also acts as the reverse primer for Hibiscus chlorotic ringspot virus (HCRSV). We have designed wet lab experiments to verify our designed primers and probes. We first checked whether the primers work appropriately. These primers should be able to amplify their target sequences, respectively. However, except for the specific primer pair, the other primers should not interfere with the target sequence. For example, primer pair LMu-03 and LMu-07 should amplify TuMV, and the other primers should not anneal to the sequence of TuMV. Fig. 3 illustrates our results with 1D gel electrophoresis. For each virus, there are two lanes, one is the amplification result of the target sequence with the presence of its specific primer pair (labeled as ‘ 2' ), the other is the amplification result with all primers presented (labeled as ‘All'). The intensities of bands on lane marked with ‘All' are usually not as strong as the bands on lane marked with ‘ 2' . For most viruses, the experimental results are ok. That is, the bands on the two lanes are identical (enclosed in solid-line squares), implying that primers did not anneal to sequences other than its own target. However, the experimental observations did not concur to the expectations for the two viruses (PVA and HCRSV, enclosed in dashed boxes). For PVA and HCRSV, the bands are not clearly visible from the gel. We have designed another experiment to verify the presence of amplified sequences. The results are illustrated in Fig. 4. We have designed slides for triplicate dot hybridization. Each slide has been sliced into 9 sections. On each region, the probe for each virus has been printed in three dots. Nine (9) slides have been prepared with identical setup. The amplified sequences were hybridized with the probes on the slide. Table 5 lists the 9 sequences, their respective primers, amplified sequence length, and the T m for each probe. The melting temperatures for the probes are high, which implies that these probes are highly specific, as we can see from Fig. 4. Most of the primers can amplify the target sequences to the amount visible from bands on the gels (Fig. 3). Some of the expected bands are not clearly observable, but can be detected by the probes in triplicate dot hybridization experiments (Fig. 4). Therefore, the integrated minimum set multiple-use primers and unique probes can indeed detect target sequences from various organisms differentially and specifically. |