Please download the zip file. Details of using this code are in the README.ranksvm file.
Authors: Ching-Pei Lee and Tzu-Ming Kuo
LIBLINEAR for more than 2^32 instances/features (experimental)
This experimental version of liblinear uses 64-bit integer in all possible places, so it in theory can handle up to 2^64 instances/features if memory is enough. Please download the zip file here. Comments are welcome.Because of converting 32-bit int to 64-bit, some warning messages may appear during compilation. Please ignore those warning message.
Author: Yu-Chin Juan
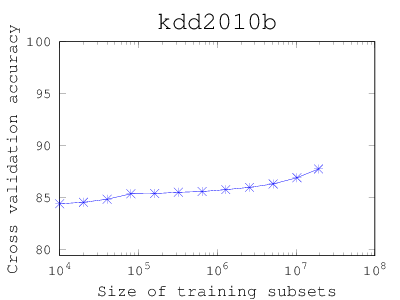
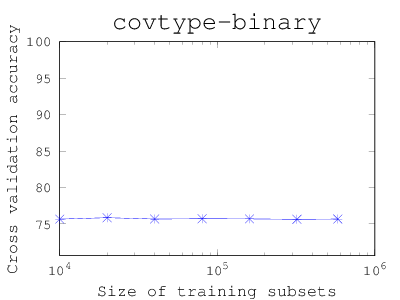
How large the training set should be?
People tend to use as many data as possible, but the training time can be long. This matlab/octave code (Download) starts with a small subset and shows if larger training subsets increase the cross-validation (CV) accuracy. The code iteratively update the figure of size versus CV accuracy. If you find that the CV accuracy has stabilized, you can stop the code and use only a subset of certain size.To use, put the code under the compiled liblinear/matlab directory, and open octave or matlab:
> [y,x] = libsvmread('mydata');
> size_acc(y,x);
Currently, only linear classification is supported
Examples:
Author: Po-Wei Wang
Large linear classification when data cannot fit in memory
This is an extension of LIBLINEAR for data which cannot fit in memory. Currently it supports L2-regularized L1- and L2-loss linear SVM, L2-regularized logistic regression, and Cramer and Singer formulation for multi-class classification problems.This code implements methods proposed in the following papers
- Hsiang-Fu Yu, Cho-Jui Hsieh, Kai-Wei Chang, and Chih-Jen Lin. Large linear classification when data cannot fit in memory, ACM Transactions on Knowledge Discovery from Data, 5:23:1--23:23, 2012. Preliminary version at ACM KDD 2010 (Best research paper award).
- Kai-Wei Chang and Dan Roth. Selective Block Minimization for Faster Convergence of Limited Memory Large-scale Linear Models, ACM KDD 2011.
Please download the zip file. Details of using this code are in the README.cdblock file. Except new parameters for this extension, the usage is the same as LIBLINEAR.
Authors: Hsiang-Fu Yu and Kai-Wei Chang
Weights for data instances
Users can give a weight to each data instance.For LIBSVM users, please download the zip file (MATLAB and Python interfaces are included).
For LIBLINEAR users, please download the zip file (MATLAB and Python interfaces are included).
- You must store weights in a separated file and specify -W your_weight_file. This setting is different from earlier versions where weights are in the first column of training data.
- Training/testing sets are the same as those for standard LIBSVM/LIBLINEAR.
- We do not support weights for test data.
- All solvers (except one-class SVM in LIBLINEAR) are supported
- Matlab/Python interfaces for both LIBSVM/LIBLIENAR are supported.
Author: Ming-Wei Chang, Hsuan-Tien Lin, Ming-Hen Tsai, Chia-Hua Ho and Hsiang-Fu Yu.
Fast training/testing for polynomial mappings of data
This is an extension of LIBLINEAR for fast training/testing of the polynomial mappings of data. Currently we support degree-2 mapping for L2-regularized L1- and L2-loss linear SVC/SVR. If you need a higher degree mapping, we also have preliminary code. Please contact us.
The implementation is baed on one method proposed in the paper
Yin-Wen Chang, Cho-Jui Hsieh, Kai-Wei Chang, Michael Ringgaard, and
Chih-Jen Lin.
Low-degree Polynomial Mappings of Data for SVM, 2009.
Please download the zip file here. Details of using this code are in the README.poly2 file. Except new parameters for the degree-2 mapping, the usage is the same as LIBLINEAR.
Authors: Yin-Wen Chang, Cho-Jui Hsieh, Kai-Wei Chang, and Yu-Chin Juan
Cross Validation with Different Criteria (AUC, F-score, etc.)
For some unbalanced data sets, accuracy may not be a good criterion for evaluating a model. This tool enables LIBSVM and LIBLINEAR to conduct cross-validation and prediction with respect to different criteria (F-score, AUC, etc.).
DetailsAuthors: Hsiang-Fu Yu, Chia-Hua Ho, Cheng-Hao Tsai, and Jui-Nan Yen
Cross Validation using Higher-level Information to Split Data
Assume you have 20,000 images of 200 users:
- User 1: 100 images
- ...
- User 200: 100 images
LIBSVM for dense data
LIBSVM stores instances as sparse vectors. For some applications, most feature values are non-zeros, so using a dense representation can significantly save the computational time. The zip file here is an implementation for dense data. See README for some comparisons with the standard libsvm.Author: Ming-Fang Weng
LIBSVM for string data
For some applications, data instances are strings. SVM trains a model using some string kernels. This experimental code (download zip file here) allows string inputs and implements one string kernel. Details are in README.string.Author: Guo-Xun Yuan
Multi-label classification
This web page contains various tools for multi-label classification.LIBSVM Extensions at Caltech
You can link to this webpage, which is individually maintained by a PhD student Hsuan-Tien Lin at Caltech. The page contains some programs that he has developed for related research. Most of these programs are extended from/for LIBSVM. Some of the most useful programs include confidence margin/decision value output, infinite ensemble learning with SVM, dense format, and MATLAB implementation for estimating posterior probability.Feature selection tool
This is a simple python script (download here) to use F-score for selecting features. To run it, please put it in the sub-directory "tools" of LIBSVM.Usage: ./fselect.py training_file [testing_file]Output files: .fscore shows importance of features, .select gives the running log, and .pred gives testing results.
More information about this implementation can be found in Y.-W. Chen and C.-J. Lin, Combining SVMs with various feature selection strategies. To appear in the book "Feature extraction, foundations and applications." 2005. This implementation is still preliminary. More comments are very welcome.
Author: Yi-Wei Chen
LIBSVM data sets
We now have a nice web page showing available data sets.SVM-toy based on Javascript
The javascript-based toy is available on LIBSVM home page. Three javascript files are included by the HTML page: libsvm-js-interfaces.js, libsvm-js-interfaces-wrappers.js, and svm-toy.js. This HTML example shows how to use these scripts. Note that libsvm-js-interfaces.js is generated by emscripten:
A. Zakai. Emscripten: an llvm-to-javascript compiler. In Proceedings of the ACM international conference companion on object oriented programming systems languages and applications companion. 2011.
If you want to generate libsvm-js-interfaces.js by yourself, in addition to the LIBSVM package, you also need js-interfaces.c and a revised Makefile. You must install emscripten, LLVM, and nodejs on your system because emscripten takes LLVM bytecode genereted from C/C++ and requires the latest version of nodejs. We provide a simple shell script get_emscripten.sh so you can put it in the desired directory and run it for automatic installation. You then must properly set
EMSCRIPTEN_ROOT = '$dir/emscripten/' LLVM_ROOT = '$dir/emscripten-fastcomp/build/Release/bin' NODE_JS = '$dir/node/node'in your ~/.emscripten, where $dir is where your install emscripten, llvm, and nodejs.
Author: Chia-Hua Ho
SVM-toy in 3D
A simple applet demonstrating SVM classification and regression in 3D. It extends the java svm-toy in the LIBSVM package.
Multi-class classification (and probability output) via error-correcting codes
Note: libsvm does support multi-class classification. The code here implements some extensions for experimental purposes.
This code implements multi-class classification and probability estimates using 4 types of error correcting codes. Details of the 4 types of ECCs and the algorithms can be found in the following paper:
T.-K. Huang, R. C. Weng, and C.-J. Lin. Generalized Bradley-Terry Models and Multi-class Probability Estimates. Journal of Machine Learning Research, 7(2006), 85-115. A (very) short version of this paper appears in NIPS 2004.
The code can be downloaded here. The installation is the same as the standard LIBSVM package, and different types of ECCs are specified as the "-i" option. Type "svm-train" without any arguments to see the usage. Note that both "one-againse-one" and "one-against-the rest" multi-class strategies are part of the implementation.
If you specify -b in training and testing, you get probability estimates and the predicted label is the one with the largest value. If you do not specify -b, this is classification based on decision values. Now we use the "exponential-loss" method in the paper:
Allwein et al.: Reducing multiclass to binary: a unifying approach for margin classifiers. Journal of Machine Learning Research, 1:113--141, 2001,
to predict class label. For one-against-the rest
(or called 1vsall), this is the same as the commonly
used way
argmax_{i} (decision value of ith class vs the rest).
For one-against-one, it is different from the
max-win strategy used in libsvm.
MATLAB code for experiments in our paper is available here
Author: Tzu-Kuo Huang
SVM Multi-class Probability Outputs
This code implements different strategies for multi-class probability estimates from in the following paperT.-F. Wu, C.-J. Lin, and R. C. Weng. Probability Estimates for Multi-class Classification by Pairwise Coupling. Journal of Machine Learning Research, 2004. A short version appears in NIPS 2003.
After libsvm 2.6, it already includes one of the methods here. You may directly use the standard libsvm unless you are interested in doing comparisons. Please download the tgz file here. The data used in the paper is available here. Please then check README for installation.
Matlab programs for the synthetic data experiment in the paper can be found in this directory. The main program is fig1a.m
Author: Tingfan Wu (svm [at] future.csie.org)
An integrated development environment to libsvm
This is a graphical environment for doing experiments with libsvm. You can create and connect components (like scaler, trainer, predictor, etc) in this environment. The program can be extended easily by writing more "plugins". It was written in python and uses wxPython library. Please download the zip file here. After unzip the package, run the file wxApp1.py. You then have to give the path of libsvm binary files in plugin/svm/svm_interface.py.
Author: Chih-Chung Chang
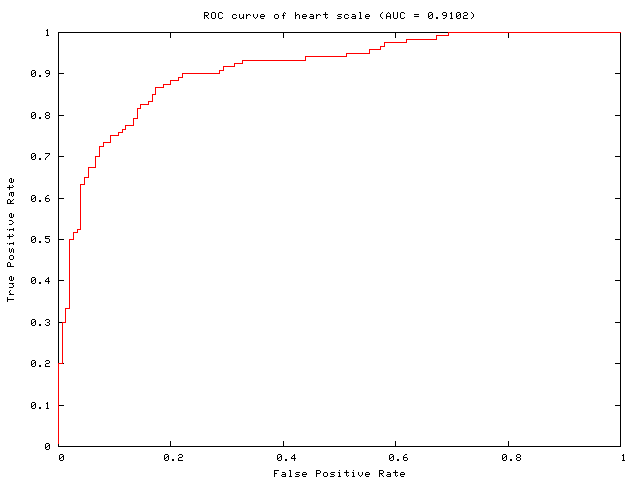
ROC Curve for Binary SVM
This tool which gives the ROC (Receiver Operating Characteristic) curve and AUC (Area Under Curve) by ranking the decision values. Note that we assume labels are +1 and -1. Multi-class is not supported yet.
You can use either MATLAB or Python.
If using MATLAB, you need to
- Download LIBSVM MATLAB interface from LIBSVM page and build it.
- Download plotroc.m to the main directory of LIBSVM MALTAB interface.
- Type
> help plotroc
If using Python, you need to
- Download LIBSVM (version 2.91 or after) and make the LIBSVM python interface.
- Download plotroc.py to the python directory.
- Edit the path of gnuplot in plotroc.py in necessary.
- The usage is
plotroc.py [-v cv_fold | -T testing_file] [libsvm_options] training_file
- Example:
> plotroc.py -v 5 -c 10 ../heart_scale
To use LIBLINEAR, you need the following modifications
- MATLAB: Copy plotroc.m to the matlab directory (note that matlab interface is included in LIBLINEAR). Replace svmtrain and svmpredict with train and predict, respectively.
Authors: Tingfan Wu (svm [at] future.csie.org), Chien-Chih Wang (d98922007 [at] ntu.edu.tw), and Hsiang-Fu Yu
Grid Parameter Search for Regression
This file is a slight modification of grid.py in the "tools" directory of libsvm. In addition to parameters C, gamma in classification, it searches for epsilon as well.
Usage: grid.py [-log2c begin,end,step] [-log2g begin,end,step] [-log2p begin,end,step] [-v fold] [-svmtrain pathname] [-gnuplot pathname] [-out pathname] [-png pathname] [additional parameters for svm-train] dataset
Author: Hsuan-Tien Lin (initial modification); Tzu-Kuo Huang (the parameter epsilon); Wei-Cheng Chang.
Radius Margin Bounds for SVM Model Selection
This is the code used in the paper: K.-M. Chung, W.-C. Kao, T. Sun, L.-L. Wang, and C.-J. Lin. Radius Margin Bounds for Support Vector Machines with the RBF Kernel. Please download the tar.bz2 file here. Details of using this code are in the readme.txt file. Part of the optimization subroutines written in Python were based on the module by Travis E. Oliphant.Author: Wei-Chun Kao with the help from Leland Wang, Kai-Min Chung, and Tony Sun
Reduced Support Vector Machines Implementation
This is the code used in the paper: K.-M. Lin and C.-J. Lin. A study on reduced support vector machines. IEEE Transactions on Neural Networks, 2003.Please download the .tgz file here. After making the binary files, type svm-train to see the usage. It includes different methods to implement RSVM.
To speed up the code, you may want to link the code to optimized BLAS/LAPACK or ATLAS.
Author: Kuan-Min Lin
LIBSVM for SVDD and finding the smallest sphere containing all data
SVDD is another type of one-class SVM proposed by Tax and Duin, Support Vector Data Description, Machine Learning, vol. 54, 2004, 45-66. Please download this zip file, put sources into libsvm-3.22 (available here), and make the code. The options are- -s 5 SVDD
- -s 6 gives the square of the radius for L1-loss SVM
- -s 7 gives the square of the radius for L2-loss SVM
For details of our SVDD formulation and implementation, please see W.-C. Chang, C.-P. Lee, and C.-J. Lin A Revisit to Support Vector Data Description (SVDD). Technical report 2013. Note that you should choose a value C in [1/l, 1], where l is the number of data. Models with C>1 are the same, and so are models with C<1/l.
For details of the square of the radius, please see K.-M. Chung, W.-C. Kao, C.-L. Sun, L.-L. Wang, and C.-J. Lin. Radius Margin Bounds for Support Vector Machines with the RBF Kernel Neural Computation, 15(2003), 2643-2681.
Authors: Leland Wang, Holger Froehlich (University of Tuebingen), Konrad Rieck (Fraunhofer institute), Chen-Tse Tsai, Tse-Ju Lin, Wei-Cheng Chang, Ching-Pei Lee
DAG approach for multiclass classification
In svm.cpp, please replace things after the first lineint nr_class = model->nr_class;in the subtoutine svm_predict() of svm.cpp with this segment of code. Note that this change causes that svm_predict() works only for classification instead of regression or one-class SVM.
This follows from the code used in the paper: C.-W. Hsu and C.-J. Lin. A comparison of methods for multi-class support vector machines , IEEE Transactions on Neural Networks